Research > Focus A > Work Packages A02
SUMMARY
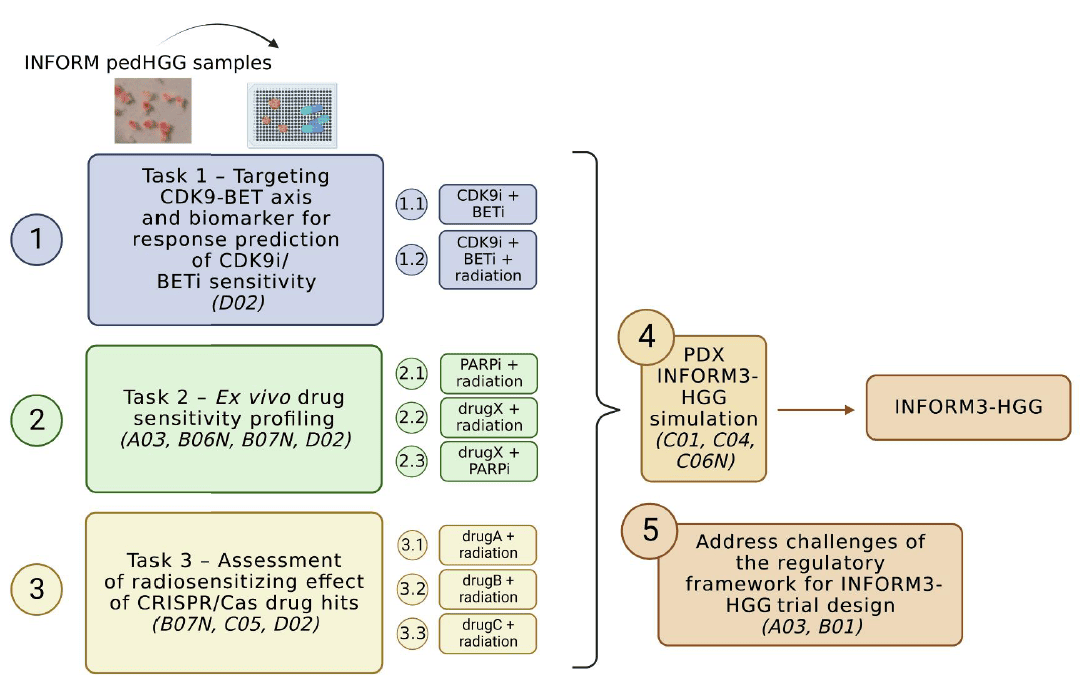
In this project we will identify a histone H3 mutation- specific combination treatment strategy for H3.3-mutant glioblastoma, which will directly feed into a three-arm umbrella trial termed INFORM3-HGG. Patient-derived pedHGG tumor samples obtained through the INFORM registry will be screened to confirm the previously identified CDK9-BET axis but also to potentially detect further novel drug efficacies in addition to the CDK9-BET axis. Furthermore, we will test top candidates from our pedHGG cell line CRISPR screens in combination with radiation modalities.
Task 1
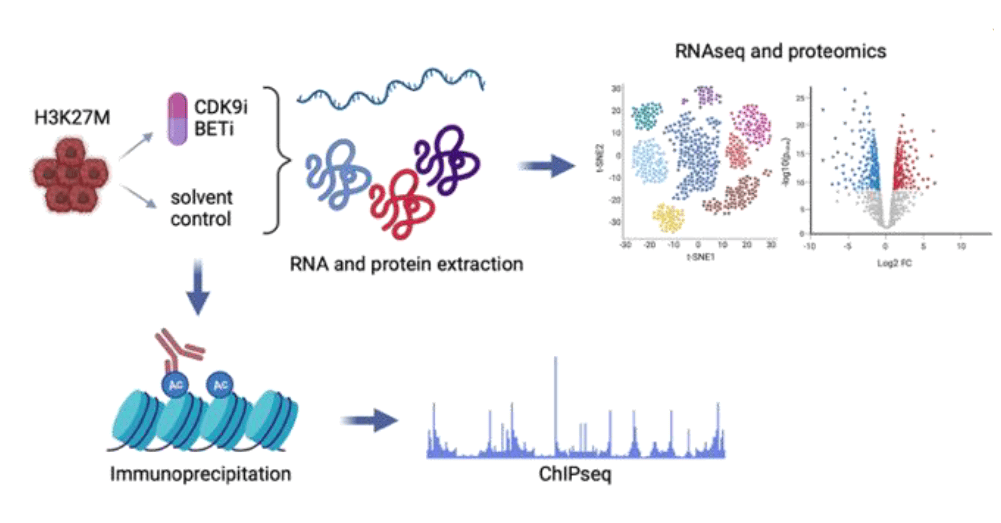
Targeting of the CDK9-BET axis and biomarker identification for drug response in H3K27M pedHGG cells. In collaboration with D02.
Steps / Workflow
- Molecular characterization of the treatment effects on H3K27M pedHGG cells
- RNAseq and proteomics analysis of treated and untreated pedHGG cells with different H3 mutation status to identify differentially expressed genes and proteins
- Assessment of the locations of oncohistones on DNA
Task 2
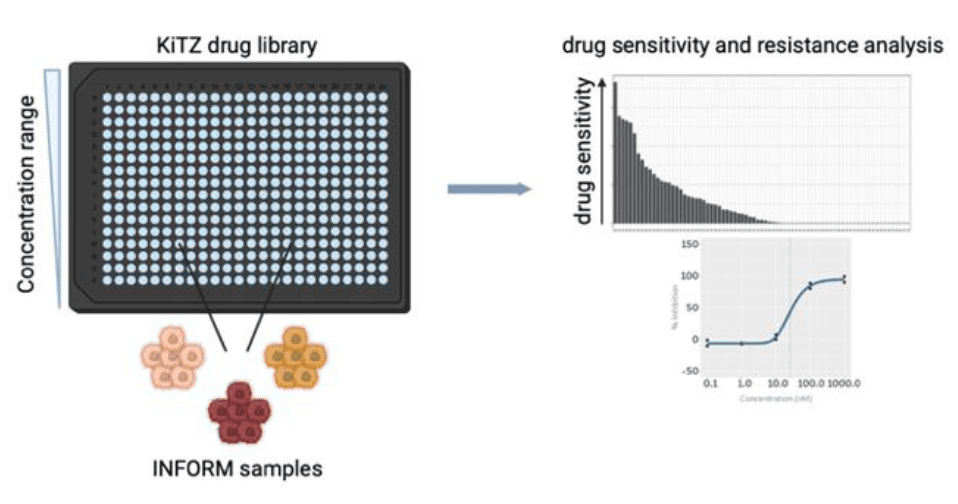
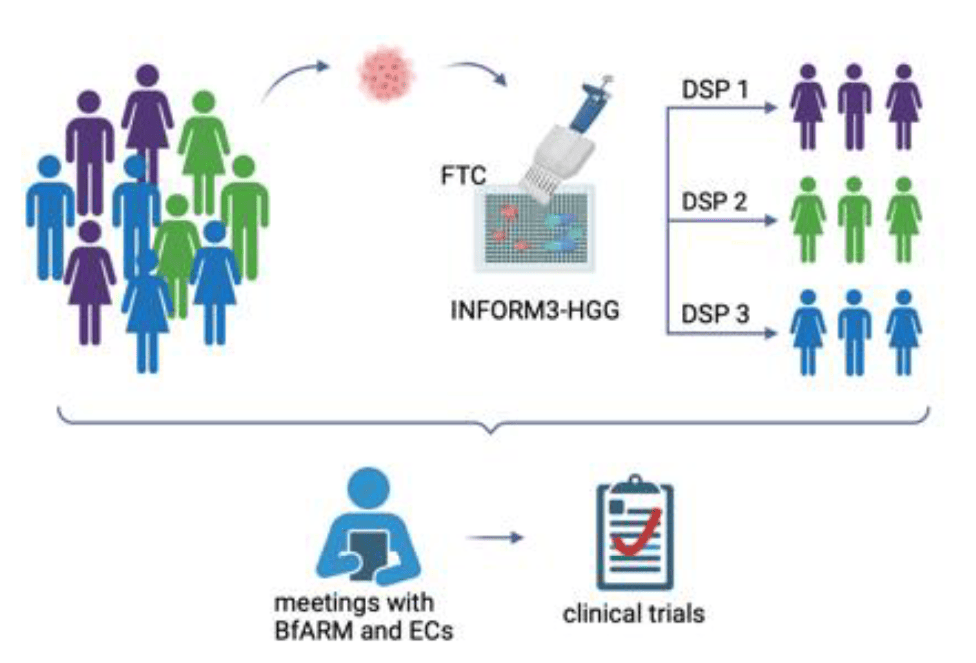
Ex vivo drug sensitivity profiling of patient HGG samples in INFORM (1, 2) with A03, B06N, B07N, D02
1 ElHarouni, Pharmacol Res. 2022; 2 Peterziel, npj Precision Oncology 2022
Steps / Workflow
- KiTZ drug library screening of INFORM samples
- Generation of dose response profiles
- Identification of top hit compounds
Task 3
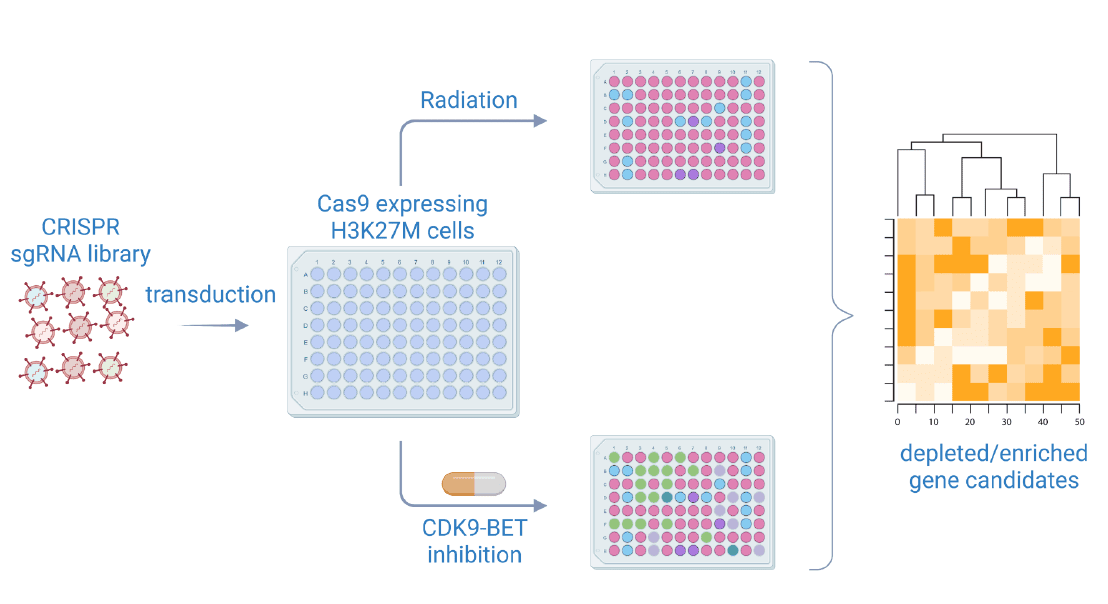
Identification of novel vulnerabilities using gene editing candidates and radiosensitizers with B07N, C05, D02
Steps / Workflow
- CRISPR/Cas mediated knockout of genes of interest
- Identification of enriched and depleted gene candidates upon simultaneous irradiation or treatment with CDK-BET inhibitor combination
- Cell culture validation of target genes
Task 4
In vivo confirmation of top hits from Task 1-3- in a zPDX umbrella simulation trial (patientderived zebrafish xenotransplantation model) with C01, C04, C06N
Steps / Workflow
- Fresh tumor tissue transplantation into zPDX (patient-derived zebrafish xenotransplantation model)
- Prioritization of drug combination efficacy in an in vivo system to inform clinical trial design (3-arm INFORM3-HGG trial)
Task 5
Address the challenges of the regulatory framework for an INFORM3-HGG trial with A03, B01
Steps / Workflow
- Ex vivo DSP to be accepted as biomarker for patientselection and enrollment into clinical trials
- Discussion with the respective competent authorities

Jones, David, Dr.
Hopp Children’s Cancer Center at the NCT Heidelberg (KiTZ) & German Cancer Research Center (DKFZ), Pediatric Glioma Research Group, Im Neuenheimer Feld 280, 69120 Heidelberg, Germany

WITT, OLAF, PROF. DR. MED.
Hopp Children’s Cancer Center at NCT Heidelberg (KiTZ) Department of Pediatric Oncology, Hematology, Immunology and Pulmonology, University Hospital Heidelberg, Im Neuenheimer Feld 430, 69120 Heidelberg, Germany
Address
Im Neuenheimer Feld 400
69120 Heidelberg
Themen
Research
- Focus A
- A01: Targeting tumor cell network communication to overcome primary and adaptive resistance in glioblastoma
- A02: Development of a specific combination therapy for histone H3-mutant pediatric glioblastoma
- A03: Deciphering resistance against targeted treatments
- A04: Elucidating tumor-associated microglia interactions in astrocytomas CNS WHO-grade 4
- A05: Predictive biomarkers for MGMT-promoter-methylated glioblastoma (2019 – 2023)
- A06: Resistance mechanisms of glioblastoma against alkylating agents and radiotherapy
- A07: Mapping and targeting neuron-tumor networks to tackle therapy resistance in glioblastoma
- A08: Personalized glioblastoma treatment guided by patient-derived tumor organoids
Research
- Focus B
- B01: Mechanisms of response and resistance to glioma-specific t cells
- B02: DNA mis-match repair regulates immune checkpoint blockade therapy in glioblastoma (2019 – 2023)
- B03: Targeting immunosuppressive programs in isocitrate dehydrogenase mutant gliomas
- B04: Impact of myeloid cells on the adaptive immune response in newly diagnosed and recurrent glioblastomas
- B05: Dissecting the response of glioblastoma and its tumor microenvironment to focused high-dose radiotherapy (2019 – 2023)
- B06: Visualization and characterization of immune responses in H3K27M mutant gliomas
Research
- Focus C
- C01: Comprehensive preclinical pharmacology testing of drugs used for glioblastoma treatment
- C02: Radiomics, radiogenomics and deep-learning in neurooncology
- C03: Imaging immune signatures of glioma response and resistance towards immunotherapy (2019 – 2023)
- C04: Metabolic signaling in glioblastoma: a spatial multi-omics approach
- C05: Overcoming glioma radio-resistance with particle therapy
- C06: Functional characterization of EGFR structural variants associated with long-term survival in glioblastoma, IDH-WT